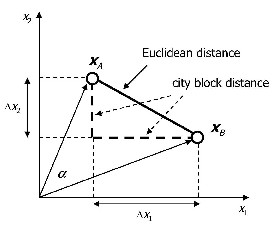
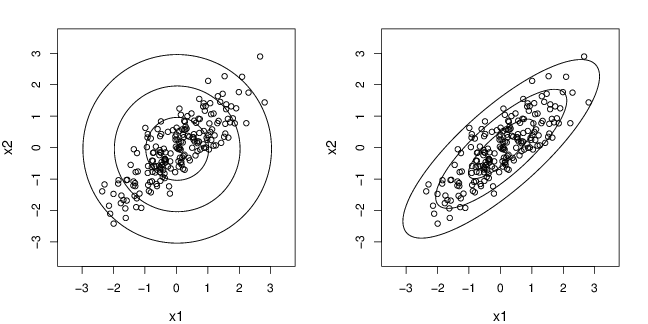
{
acov <- var(a)
cov.svd <- svd(acov, nv = 0)
me <- apply(a, 2, mean)
acov <- var(a)
cov.svd <- svd(acov, nv = 0)
r <- cov.svd[["u"]] %*% diag(sqrt(cov.svd[["d"]]))
m <- 1000
alphaed <- c(3,2,1)
alphamd <- c(3,2,1)
par(pty="s")
lalpha <- length(alphaed)
for(j in 1:lalpha) {
if (which=="MD"){
e1md <- cos(c(0:m)/m * 2 * pi) * alphamd[j]
e2md <- sin(c(0:m)/m * 2 * pi) * alphamd[j]
emd <- cbind(e1md, e2md)
ttmd <- t(r %*% t(emd)) + rep(1, m + 1) %o% me
lines(ttmd[, 1], ttmd[, 2], ...)
}
else {
e1ed <- cos(c(0:m)/m * 2 * pi) * alphaed[j]
e2ed <- sin(c(0:m)/m * 2 * pi) * alphaed[j]
eed <- cbind(e1ed, e2ed)
tted <- t(t(eed)) + rep(1, m + 1) %o% me
lines(tted[, 1], tted[, 2], ...)
}
}
}
pdf("mahalanobis.pdf",width=9,height=4.5)
par(mfrow=c(1,2))
par(mar=c(4,4,2,2))
library(mvtnorm)
set.seed(102)
x=rmvnorm(200,c(0,0),matrix(c(1,0.8,0.8,1),ncol=2))
plot(x,xlab="x1",ylab="x2",cex.lab=1.2,xlim=c(-3.5,3.5),ylim=c(-3.5,3.5))
do_elipses (x, which="ED", type = "l",col=1,lty=1,lwd=1)
plot(x,xlab="x1",ylab="x2",cex.lab=1.2,xlim=c(-3.5,3.5),ylim=c(-3.5,3.5))
do_elipses (x, which="MD", type = "l",col=1,lty=1,lwd=1)
dev.off()