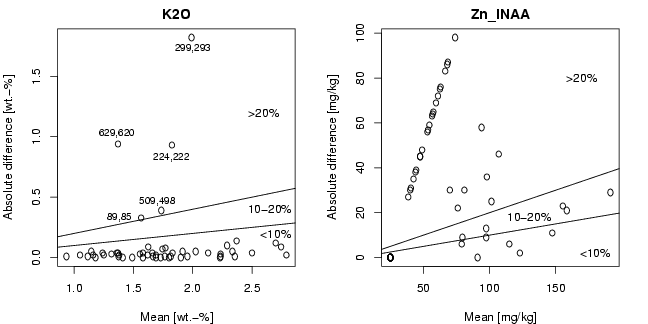
library(StatDA)
# Analytical duplicates
data(CHorANADUP)
xd1=CHorANADUP[,3:96]
xd2=CHorANADUP[,97:190]
pdf("fig-18-2.pdf",width=9,height=4.5)
par(mfrow=c(1,2),mar=c(4,4,2,2))
i=39
xmean=apply(cbind(xd1[,i],xd2[,i]),1,mean)
xdiff=abs(xd1[,i]-xd2[,i])
xmax=max(xmean)
ymax=max(xdiff)
plot(xmean,xdiff,main="K2O",xlab="Mean [wt.-%]",ylab="Absolute difference [wt.-%]",
ylim=c(0,ymax))
abline(a=0,b=0.1)
abline(a=0,b=0.2)
text(2.7,0.2,"<10%")
text(2.65,0.4,"10-20%")
text(2.6,1.2,">20%")
# label observations
text(1.38,0.33,"89,85",cex=0.85)
text(1.82,0.83,"224,222",cex=0.85)
text(2.0,1.73,"299,293",cex=0.85)
text(1.7,0.47,"509,498",cex=0.85)
text(1.36,1.03,"629,620",cex=0.85)
i=94
xmean=apply(cbind(xd1[,i],xd2[,i]),1,mean)
xdiff=abs(xd1[,i]-xd2[,i])
xmax=max(xmean)
ymax=max(xdiff)
plot(xmean,xdiff,main="Zn_INAA",xlab="Mean [mg/kg]",ylab="Absolute difference [mg/kg]",
ylim=c(0,ymax))
abline(a=0,b=0.1)
abline(a=0,b=0.2)
text(180,2,"<10%")
text(130,18,"10-20%")
text(170,80,">20%")
dev.off()