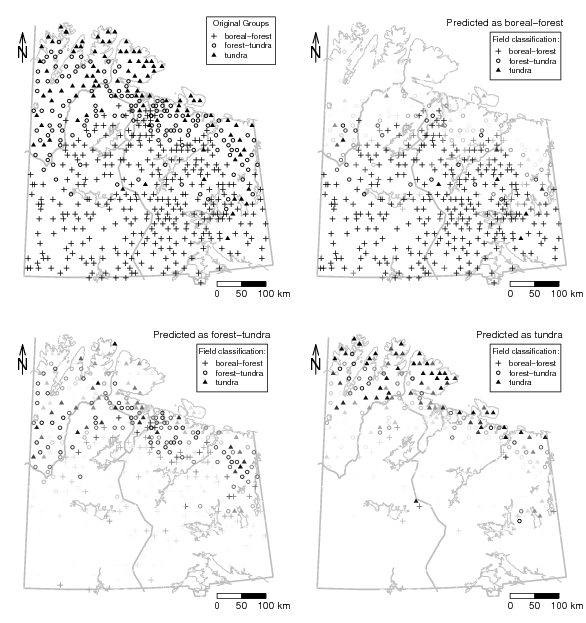
library(StatDA)
data(kola.background)
# DA with vegetation zones
data(ohorizon)
vegzn=ohorizon[,"VEG_ZONE"]
veg=rep(NA,nrow(ohorizon))
veg[vegzn=="BOREAL_FOREST"] <- 1
veg[vegzn=="FOREST_TUNDRA"] <- 2
veg[vegzn=="SHRUB_TUNDRA"] <- 3
veg[vegzn=="DWARF_SHRUB_TUNDRA"] <- 3
veg[vegzn=="TUNDRA"] <- 3
#el=c("Ag","B","Bi","Ca_AA","Cd","Cu","Fe_AA","K_AA","Mg_AA","Mn_AA","Pb")
#el=c("Ag","B","Bi","Ca_AA","Cd","Cu","Fe_AA","K_AA","Mg_AA","Mn_AA","Pb")
#el=c("Al_AA","Ba_AA","Ca_AA","Co_AA","Cu_AA","Fe_AA","K_AA","Mg_AA","Mn_AA",
# "Ni_AA","S_AA","SR_AA","Zn_AA")
el=c("Ag","Al","As","B","Ba","Bi","Ca","Cd","Co","Cu","Fe","K","Mg","Mn",
"Na","Ni","P","Pb","Rb","S","Sb","Sr","Th","Tl","V","Y","Zn")
##el=c("Ag","Al","As","Ba","Bi","Ca","Cd","Co","Cu","Fe","K","Mn",
## "Ni","P","Pb","Rb","Sb","Th","Tl","V","Y","Zn")
x <- log10(ohorizon[!is.na(veg),el])
#x <- log10(ohorizon[!is.na(veg),c(14:17,19:32,34:61,64,66:85)])
#x=scale(x)
v <- veg[!is.na(veg)]
X=ohorizon[!is.na(veg),"XCOO"]
Y=ohorizon[!is.na(veg),"YCOO"]
XY=cbind(X,Y)
# true representation of x and y axis of map for plot
xwid=diff(range(X))/12e4
ywid=diff(range(Y))/12e4
pdf("fig-17-3.pdf",width=2*xwid,height=2*ywid)
par(mfrow=c(2,2),mar=c(1.5,1.5,1.5,1.5))
# generate plot with background
plot(X,Y,frame.plot=FALSE,xaxt="n",yaxt="n",xlab="",ylab="",type="n")
plotbg(map.col=c("gray","gray","gray","gray"),add.plot=T)
points(XY[v==1,],pch=3,cex=0.8)
points(XY[v==2,],pch=1,cex=0.8)
points(XY[v==3,],pch=17,cex=0.8)
set.seed(201)
#x.rqda=qda(x, v, subset=train, method="mve")
#x.rqda=lda(x, v, subset=train)
x.rqda=lda(x, v, CV=TRUE)
#x.rqda.pred=predict(x.rqda,x)
cl=x.rqda$class
pt=x.rqda$post
pt.na=apply(is.na(pt),1,sum)
cl.red=cl[pt.na==0]
pt.red=pt[pt.na==0,]
legend("topright",pch=c(3,1,17),pt.cex=c(0.8,0.8,0.8),
legend=c("boreal-forest","forest-tundra","tundra"),
cex=0.8,title="Original Groups")
# scalebar
scalebar(761309,7373050,861309,7363050,shifttext=-0.5,shiftkm=37e3,sizetext=0.8)
# North arrow
Northarrow(362602,7818750,362602,7878750,362602,7838750,Alength=0.15,Aangle=15,Alwd=1.3,Tcex=1.6)
###################################################################################
# Results Group 1
# generate plot with background
plot(X,Y,frame.plot=FALSE,xaxt="n",yaxt="n",xlab="",ylab="",type="n")
plotbg(map.col=c("gray","gray","gray","gray"),add.plot=T)
#points(XY,pch=15,cex=0.6,col=gray(1-pt.red[,1]))
v.red=v[pt.na==0]
XY.red=XY[pt.na==0,]
points(XY.red[v.red==1,],pch=3,cex=0.8,col=gray(1-pt.red[v.red==1,1]))
points(XY.red[v.red==2,],pch=1,cex=0.8,col=gray(1-pt.red[v.red==2,1]))
points(XY.red[v.red==3,],pch=17,cex=0.8,col=gray(1-pt.red[v.red==3,1]))
#points(XY[cl.red==1,],pch=15,cex=0.6,col=gray(1-pt.red[cl.red==1,1]))
#points(XY[cl.red==2,],pch=1,cex=0.6,col=gray(1-pt.red[cl.red==2,2]))
#points(XY[cl.red==3,],pch=17,cex=0.6,col=gray(1-pt.red[cl.red==3,3]))
text(750000,7900000,"Predicted as boreal-forest",cex=1)
legend(720000,7880000,pch=c(3,1,17),pt.cex=c(0.8,0.8,0.8),
legend=c("boreal-forest","forest-tundra","tundra"),
cex=0.8,title="Field classification:")
# scalebar
scalebar(761309,7373050,861309,7363050,shifttext=-0.5,shiftkm=37e3,sizetext=0.8)
# North arrow
Northarrow(362602,7818750,362602,7878750,362602,7838750,Alength=0.15,Aangle=15,Alwd=1.3,Tcex=1.6)
###################################################################################
# Results Group 2
# generate plot with background
plot(X,Y,frame.plot=FALSE,xaxt="n",yaxt="n",xlab="",ylab="",type="n")
plotbg(map.col=c("gray","gray","gray","gray"),add.plot=T)
#points(XY,pch=15,cex=0.6,col=gray(1-pt.red[,1]))
v.red=v[pt.na==0]
XY.red=XY[pt.na==0,]
points(XY.red[v.red==1,],pch=3,cex=0.8,col=gray(1-pt.red[v.red==1,2]))
points(XY.red[v.red==2,],pch=1,cex=0.8,col=gray(1-pt.red[v.red==2,2]))
points(XY.red[v.red==3,],pch=17,cex=0.8,col=gray(1-pt.red[v.red==3,2]))
#points(XY[cl.red==1,],pch=15,cex=0.6,col=gray(1-pt.red[cl.red==1,1]))
#points(XY[cl.red==2,],pch=1,cex=0.6,col=gray(1-pt.red[cl.red==2,2]))
#points(XY[cl.red==3,],pch=17,cex=0.6,col=gray(1-pt.red[cl.red==3,3]))
text(750000,7900000,"Predicted as forest-tundra",cex=1)
legend(720000,7880000,pch=c(3,1,17),pt.cex=c(0.8,0.8,0.8),
legend=c("boreal-forest","forest-tundra","tundra"),
cex=0.8,title="Field classification:")
# scalebar
scalebar(761309,7373050,861309,7363050,shifttext=-0.5,shiftkm=37e3,sizetext=0.8)
# North arrow
Northarrow(362602,7818750,362602,7878750,362602,7838750,Alength=0.15,Aangle=15,Alwd=1.3,Tcex=1.6)
###################################################################################
# Results Group 3
# generate plot with background
plot(X,Y,frame.plot=FALSE,xaxt="n",yaxt="n",xlab="",ylab="",type="n")
plotbg(map.col=c("gray","gray","gray","gray"),add.plot=T)
#points(XY,pch=15,cex=0.6,col=gray(1-pt.red[,1]))
v.red=v[pt.na==0]
XY.red=XY[pt.na==0,]
points(XY.red[v.red==1,],pch=3,cex=0.8,col=gray(1-pt.red[v.red==1,3]))
points(XY.red[v.red==2,],pch=1,cex=0.8,col=gray(1-pt.red[v.red==2,3]))
points(XY.red[v.red==3,],pch=17,cex=0.8,col=gray(1-pt.red[v.red==3,3]))
#points(XY[cl.red==1,],pch=15,cex=0.6,col=gray(1-pt.red[cl.red==1,1]))
#points(XY[cl.red==2,],pch=1,cex=0.6,col=gray(1-pt.red[cl.red==2,2]))
#points(XY[cl.red==3,],pch=17,cex=0.6,col=gray(1-pt.red[cl.red==3,3]))
text(780000,7900000,"Predicted as tundra",cex=1)
legend(720000,7880000,pch=c(3,1,17),pt.cex=c(0.8,0.8,0.8),
legend=c("boreal-forest","forest-tundra","tundra"),
cex=0.8,title="Field classification:")
# scalebar
scalebar(761309,7373050,861309,7363050,shifttext=-0.5,shiftkm=37e3,sizetext=0.8)
# North arrow
Northarrow(362602,7818750,362602,7878750,362602,7838750,Alength=0.15,Aangle=15,Alwd=1.3,Tcex=1.6)
###################################################################################
a1=sum(cl[v==1]!=v[v==1])
a2=sum(cl[v==2]!=v[v==2])
a3=sum(cl[v==3]!=v[v==3])
b1=a1/sum(v==1)*100
b2=a2/sum(v==2)*100
b3=a3/sum(v==3)*100
print(c(b1,b2,b3,(a1+a2+a3)/length(v)*100))
dev.off()
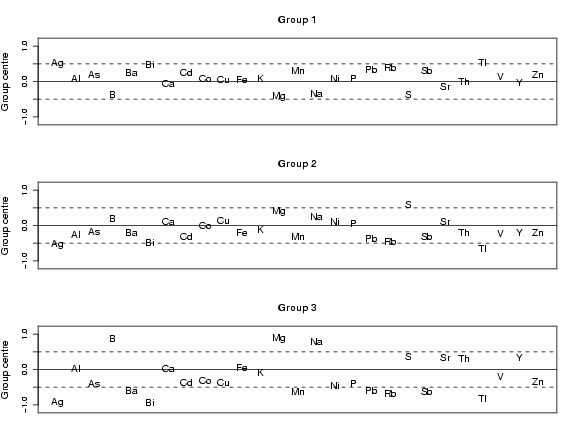
library(StatDA)
# DA with vegetation zones
data(ohorizon)
vegzn=ohorizon[,"VEG_ZONE"]
veg=rep(NA,nrow(ohorizon))
veg[vegzn=="BOREAL_FOREST"] <- 1
veg[vegzn=="FOREST_TUNDRA"] <- 2
veg[vegzn=="SHRUB_TUNDRA"] <- 3
veg[vegzn=="DWARF_SHRUB_TUNDRA"] <- 3
veg[vegzn=="TUNDRA"] <- 3
el=c("Ag","Al","As","B","Ba","Bi","Ca","Cd","Co","Cu","Fe","K","Mg","Mn",
"Na","Ni","P","Pb","Rb","S","Sb","Sr","Th","Tl","V","Y","Zn")
x <- log10(ohorizon[!is.na(veg),el])
x=scale(x)
v <- veg[!is.na(veg)]
pdf("fig-17-4.pdf",width=8,height=6)
par(mfrow=c(1,1),mar=c(2,2,2,2))
set.seed(201)
#x.rqda=qda(x, v, subset=train, method="mve")
#x.rqda=lda(x, v, subset=train)
x.da=lda(x, v)
x.da.pred=predict(x.da,x)
cl=x.da.pred$cl
plotelement(x.da)
###################################################################################
a1=sum(cl[v==1]!=v[v==1])
a2=sum(cl[v==2]!=v[v==2])
a3=sum(cl[v==3]!=v[v==3])
b1=a1/sum(v==1)*100
b2=a2/sum(v==2)*100
b3=a3/sum(v==3)*100
print(c(b1,b2,b3,(a1+a2+a3)/length(v)*100))
dev.off()