library(StatDA)
data(ohorizon)
attach(ohorizon)
sel <- c("Ag","Al","As","B","Ba","Be","Bi","Ca","Cd","Co","Cr","Cu","Fe","Hg","K","La","Mg","Mn","Mo","Na",
"Ni","P","Pb","Rb","S","Sb","Sc","Si","Sr","Th","Tl","U","V","Y","Zn","C","H","N","pH")
ohorizon.red <- ohorizon[,sel]
ohorizon.red <- na.omit(ohorizon.red)
# full model using 38 variables:
lm.full=lm(log(Bi)~log(Ag)+log(Al)+log(As)+log(B)+log(Ba)+log(Be)+log(Ca)+log(Cd)+
log(Co)+log(Cr)+log(Cu)+log(Fe)+log(Hg)+log(K)+log(La)+log(Mg)+log(Mn)+log(Mo)+log(Na)+
log(Ni)+log(P)+log(Pb)+log(Rb)+log(S)+log(Sb)+log(Sc)+log(Si)+log(Sr)+log(Th)+log(Tl)+
log(U)+log(V)+log(Y)+log(Zn)+C+H+N+pH,data=ohorizon.red)
summary(lm.full)
# empty model:
lm.0=lm(log(Bi)~1,data=ohorizon.red)
# stepwise regressions:
#backward
step.b=step(lm.full,data=ohorizon.red,direction="backward")
summary(step.b)
#forward
step.f=step(lm.0,scope=lm.full$call$formula,data=ohorizon.red,direction="forward")
summary(step.f)
#both directions
step.both=step(lm.full,direction="both")
summary(step.both)
# leaps and bound:
res=regsubsets(log(Bi)~log(Ag)+log(Al)+log(As)+log(B)+log(Ba)+log(Be)+log(Ca)+log(Cd)+
log(Co)+log(Cr)+log(Cu)+log(Fe)+log(Hg)+log(K)+log(La)+log(Mg)+log(Mn)+log(Mo)+log(Na)+
log(Ni)+log(P)+log(Pb)+log(Rb)+log(S)+log(Sb)+log(Sc)+log(Si)+log(Sr)+log(Th)+log(Tl)+
log(U)+log(V)+log(Y)+log(Zn)+C+H+N+pH,data=ohorizon.red,
nbest=1,nvmax=9)
# plot result:
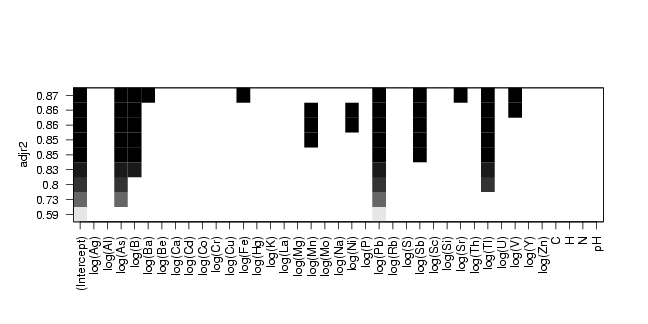
pdf("fig-16-10.pdf",width=9,height=4.5)
plot(res,scale="adjr2")
dev.off()
# linear model with best 4 variables: classical
lm.new=lm(log(Bi)~log(Pb)+log(As)+log(Tl)+log(B),data=ohorizon)
summary(lm.new)
# linear model with best 4 variables: robust
lts.new=ltsReg(log(Bi)~log(Pb)+log(As)+log(Tl)+log(B),data=ohorizon)
summary(lts.new)
ltsReg.formula(formula = log(Bi) ~ log(Pb) + log(As) + log(Tl) +
log(B), data = ohorizon.red)
Residuals (from reweighted LS):
Min 1Q Median 3Q Max
-0.34381 -0.08645 0.00000 0.10007 0.37935
Coefficients:
Estimate Std. Error t value Pr(>|t|)
Intercept -3.71126 0.10838 -34.244 < 2e-16
log(Pb) 0.77057 0.02472 31.171 < 2e-16
log(As) 0.26162 0.01404 18.635 < 2e-16
log(Tl) 0.14799 0.01727 8.571 < 2e-16
log(B) -0.11859 0.01627 -7.290 1.01e-12
Residual standard error: 0.1511 on 585 degrees of freedom
Multiple R-Squared: 0.908, Adjusted R-squared: 0.9073
F-statistic: 1443 on 4 and 585 DF, p-value: < 2.2e-16
library(StatDA)
data(ohorizon)
attach(ohorizon)
sel <- c("Ag","Al","As","B","Ba","Be","Bi","Ca","Cd","Co","Cr","Cu","Fe","Hg","K","La","Mg","Mn","Mo","Na",
"Ni","P","Pb","Rb","S","Sb","Sc","Si","Sr","Th","Tl","U","V","Y","Zn","C","H","N","pH")
ohorizon.red <- ohorizon[,sel]
ohorizon.red <- na.omit(ohorizon.red)
# full model using 38 variables:
lm.full=lm(log(Bi)~log(Ag)+log(Al)+log(As)+log(B)+log(Ba)+log(Be)+log(Ca)+log(Cd)+
log(Co)+log(Cr)+log(Cu)+log(Fe)+log(Hg)+log(K)+log(La)+log(Mg)+log(Mn)+log(Mo)+log(Na)+
log(Ni)+log(P)+log(Pb)+log(Rb)+log(S)+log(Sb)+log(Sc)+log(Si)+log(Sr)+log(Th)+log(Tl)+
log(U)+log(V)+log(Y)+log(Zn)+C+H+N+pH,data=ohorizon.red)
summary(lm.full)
# empty model:
lm.0=lm(log(Bi)~1,data=ohorizon.red)
# stepwise regressions:
#backward
step.b=step(lm.full,data=ohorizon.red,direction="backward")
summary(step.b)
#forward
step.f=step(lm.0,scope=lm.full$call$formula,data=ohorizon.red,direction="forward")
summary(step.f)
#both directions
step.both=step(lm.full,direction="both")
summary(step.both)
# leaps and bound:
res=regsubsets(log(Bi)~log(Ag)+log(Al)+log(As)+log(B)+log(Ba)+log(Be)+log(Ca)+log(Cd)+
log(Co)+log(Cr)+log(Cu)+log(Fe)+log(Hg)+log(K)+log(La)+log(Mg)+log(Mn)+log(Mo)+log(Na)+
log(Ni)+log(P)+log(Pb)+log(Rb)+log(S)+log(Sb)+log(Sc)+log(Si)+log(Sr)+log(Th)+log(Tl)+
log(U)+log(V)+log(Y)+log(Zn)+C+H+N+pH,data=ohorizon.red,
nbest=1,nvmax=9)
# plot result:
plot(res,scale="adjr2")
# linear model with best 4 variables: classical
lm.new=lm(log(Bi)~log(Pb)+log(As)+log(Tl)+log(B),data=ohorizon.red)
summary(lm.new)
# linear model with best 4 variables: robust
lts.new=ltsReg(log(Bi)~log(Pb)+log(As)+log(Tl)+log(B),data=ohorizon.red)
summary(lts.new)
sink("tab-16-4.txt")
print(summary(lts.new))
sink()