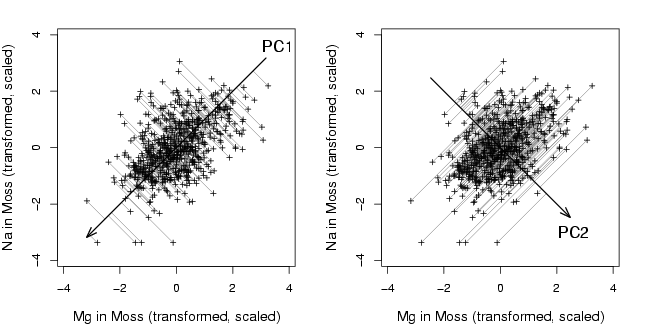
library(StatDA)
data(moss)
x=scale(log10(moss[,c("Mg","Na")]))
res=princomp(x)
loa=res$loa
sco=res$sco
pdf("fig-14-2.pdf",width=9,height=4.5)
par(mfcol=c(1,2),mar=c(4,4,2,2))
plot(x,xlab="Mg in Moss (transformed, scaled)", ylab="Na in Moss (transformed, scaled)",
cex.lab=1.2, cex=0.5, pch=3,type="n",xlim=c(-3.9,3.9),ylim=c(-3.9,3.9))
hlp <- sco[,1]%*%t(loa[,1])
for (i in 1:nrow(x)){
segments(x[i,1],x[i,2],hlp[i,1],hlp[i,2],col=gray(0.4),lwd=0.1,lty=1)
}
points(x,cex=0.7, pch=3)
arrows(-4.5*loa[1,1],-4.5*loa[2,1],4.5*loa[1,1],4.5*loa[2,1],lwd=1.5,
angle=20,length=0.15)
text(3.6,3.6,"PC1",cex=1.3)
plot(x,xlab="Mg in Moss (transformed, scaled)", ylab="Na in Moss (transformed, scaled)",
cex.lab=1.2, cex=0.5, pch=3,type="n",xlim=c(-3.9,3.9),ylim=c(-3.9,3.9))
hlp <- sco[,2]%*%t(loa[,2])
for (i in 1:nrow(x)){
segments(x[i,1],x[i,2],hlp[i,1],hlp[i,2],col=gray(0.4),lwd=0.1,lty=1)
}
points(x,cex=0.7, pch=3)
arrows(-3.5*loa[1,2],-3.5*loa[2,2],3.5*loa[1,2],3.5*loa[2,2],lwd=1.5,
angle=20,length=0.15)
text(2.6,-3.0,"PC2",cex=1.3)
dev.off()