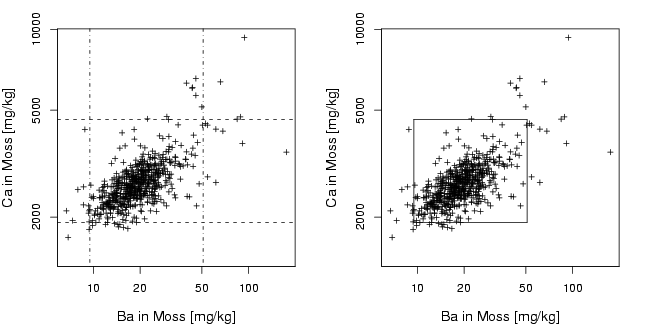
library(StatDA)
data(moss)
Ba=log10(moss[,"Ba"])
Ca=log10(moss[,"Ca"])
pdf("fig-13-1.pdf",width=9,height=4.5)
par(mfcol=c(1,2),mar=c(4,4,2,2))
plot(Ba,Ca,xlab="Ba in Moss [mg/kg]",ylab="Ca in Moss [mg/kg]",
cex.lab=1.2, cex=0.7, pch=3, xaxt="n", yaxt="n",
ylim=c(3.15,max(Ca)))
axis(1,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
axis(2,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
abline(v=quantile(Ba,c(0.02,0.98)),lty=4)
abline(h=quantile(Ca,c(0.02,0.98)),lty=2)
plot(Ba,Ca,xlab="Ba in Moss [mg/kg]",ylab="Ca in Moss [mg/kg]",
cex.lab=1.2, cex=0.7, pch=3, xaxt="n", yaxt="n",
ylim=c(3.15,max(Ca)))
axis(1,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
axis(2,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
segments(quantile(Ba,0.02),quantile(Ca,0.02),quantile(Ba,0.98),quantile(Ca,0.02))
segments(quantile(Ba,0.98),quantile(Ca,0.02),quantile(Ba,0.98),quantile(Ca,0.98))
segments(quantile(Ba,0.02),quantile(Ca,0.98),quantile(Ba,0.98),quantile(Ca,0.98))
segments(quantile(Ba,0.02),quantile(Ca,0.02),quantile(Ba,0.02),quantile(Ca,0.98))
dev.off()
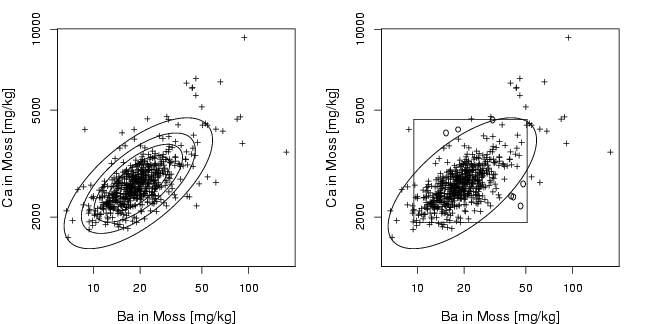
library(StatDA)
data(moss)
Ba=log10(moss[,"Ba"])
Ca=log10(moss[,"Ca"])
pdf("fig-13-2.pdf",width=9,height=4.5)
par(mfcol=c(1,2),mar=c(4,4,2,2))
plot(Ba,Ca,xlab="Ba in Moss [mg/kg]",ylab="Ca in Moss [mg/kg]",
cex.lab=1.2, cex=0.7, pch=3, xaxt="n", yaxt="n",
ylim=c(3.15,max(Ca)))
axis(1,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
axis(2,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
x=cbind(Ba,Ca)
plotellipse(apply(x,2,mean),cov(x),perc=c(0.5,0.75,0.9,0.98))
plot(Ba,Ca,xlab="Ba in Moss [mg/kg]",ylab="Ca in Moss [mg/kg]",
cex.lab=1.2, cex=0.7, pch=3, xaxt="n", yaxt="n",type="n",
ylim=c(3.15,max(Ca)))
axis(1,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
axis(2,at=log10(alog<-sort(c((10^(-50:50))%*%t(c(2,5,10))))),labels=alog)
x=cbind(Ba,Ca)
plotellipse(apply(x,2,mean),cov(x),perc=c(0.98))
segments(quantile(Ba,0.02),quantile(Ca,0.02),quantile(Ba,0.98),quantile(Ca,0.02))
segments(quantile(Ba,0.98),quantile(Ca,0.02),quantile(Ba,0.98),quantile(Ca,0.98))
segments(quantile(Ba,0.02),quantile(Ca,0.98),quantile(Ba,0.98),quantile(Ca,0.98))
segments(quantile(Ba,0.02),quantile(Ca,0.02),quantile(Ba,0.02),quantile(Ca,0.98))
# which data points outside ellipse but inside rectangular?
out.el=(mahalanobis(x,apply(x,2,mean),cov(x)) > qchisq(0.98,2))
in.rec=(Ba>quantile(Ba,0.02) & Ba
points(Ba[out.in],Ca[out.in],cex=0.9, pch=1)
points(Ba[!out.in],Ca[!out.in],cex=0.7, pch=3)
dev.off()